
methodologies
Sampling

Sample site selection
We are aiming to sample as broad a cross section of physicochemical conditions presented by geothermally-influenced springs within the Taupō Volcanic Zone as possible. In this research, we sample only the water columns of springs and not the sediments, biofilms, mudpots or heated-soils. We are aiming to collect 1000 samples which in the majority of cases are from individual features. However, in some cases, we have repeated sampling of the same feature where physicochemical conditions are known to vary, are being targeted for a longer term temporal study or for quality control reasons.
Sampling
We collect water from individual springs using the same technique for every sample. Spring water is collected across the full depth of the water column to ensure that a homogenous and representative sample. Water is collected using a telescopic sampling pole equipped with either a sterile 500 mL PP container or a custom-built sampler. We use the custom-built sampler to collect water where springs are deep enough. The sampler collects water without mixing the water column. Sampled water is then transferred to various containers for downstream microbiological and physicochemical analysis; 2150 mL of water is collected in a sterile 2 L PP container for diversity and some chemistry analysis; ~ 330 mL of water is collected in a rubber-necked bottle for hydrogen sulphide (H2S), bicarbonate (HCO3) and chloride (Cl) analysis; 500 mL of water is collected in a sterile 500 mL PP bottle for physical attributes such as conductivity and dissolved oxygen (dO) measurement; and ~60 mL of water is collected for dissolved gas content. Explanations of these analyses are given below. In addition, an additional sediment sample (~10 g) is taken where possible for future research.
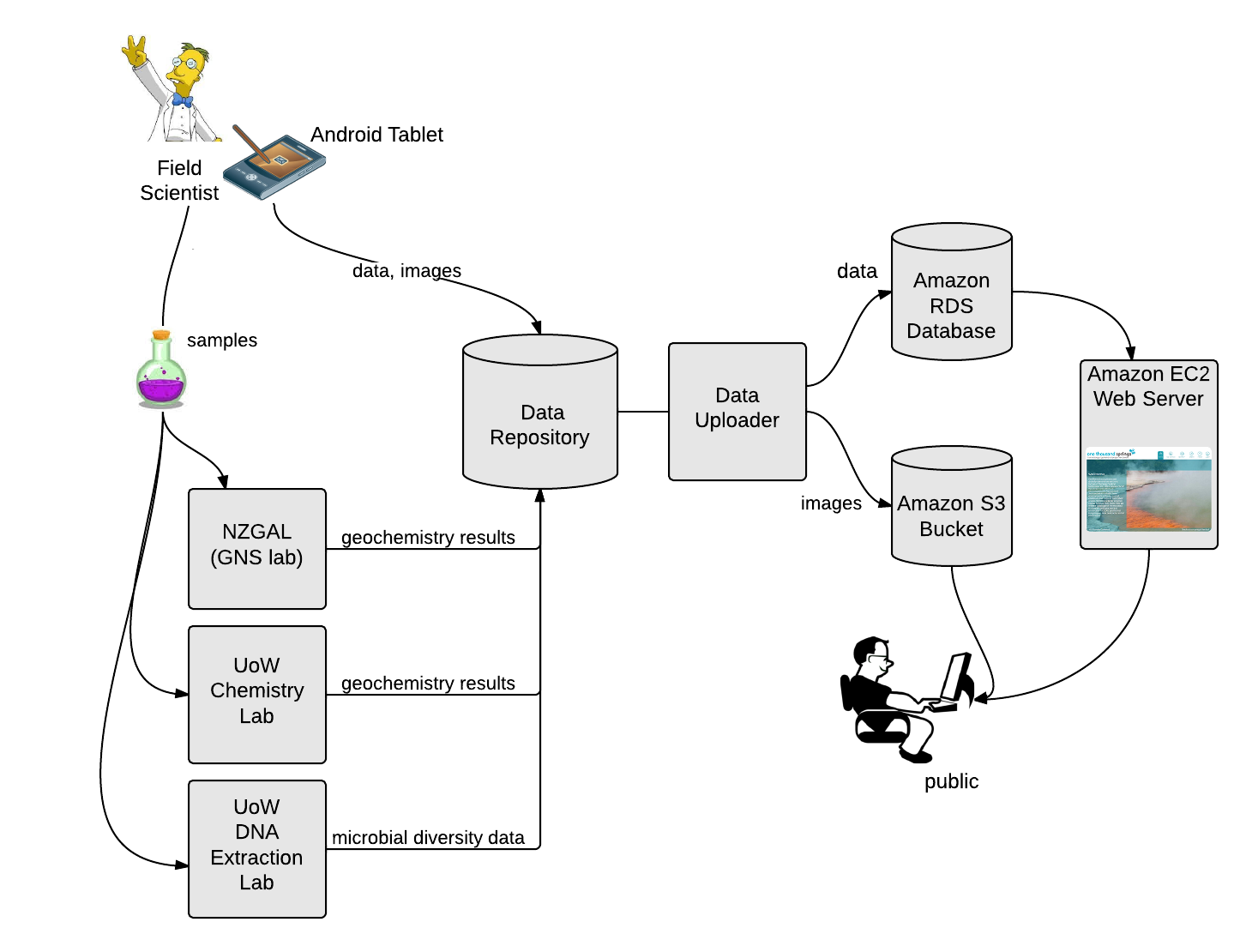
As part of this project we have developed a field tablet (see image) and a custom-built app to allow us to record physical, chemical and other metadata in the field. This avoids having to double-handle notes scribbled in the field and reduces the number of transcribing errors. If you’d like to know more about the field tablet and associated software, please contact us. The physical and chemical data we generate in the field are described elsewhere link. The metadata we record are the feature type (spring, stream etc.), area description, sampling date and sampling staff, feature location, GPS location, spring size, ebullition (bubbling/water up flow), and spring colour. We also take images and draw a ‘bird’s-eye-view’ of the spring to facilitate resampling in the future using the tablet.

Field Processing Of Samples

We aim to process samples as soon as possible to minimise any effects due to temperature or dissolved oxygen changes, or microbial activity. Samples to our 4WD field van which has been set up as a mobile laboratory. A field multiparameter meter (Hanna) is calibrated and used to measure pH, oxidation-reduction potential (ORP or redox), conductivity (a proxy for salinity), total dissolved solids, turbidity and dissolved oxygen from the (tightly-sealed) PP 500 mL bottle. Ferrous iron is immediately measured using a field spectrophotometer (Wilson, 1960).
Microorganisms are collected via filtration. A battery operated peristaltic pump pushes up to 2 L of sample through a 0.2 µm Sterivex column filter (Millipore). After a lot of field testing we found that Sterivex filters had a higher filtration volume capacity compared to all other filter types we tested. Filtrate water was collected in 15 mL and 50 mL tubes for other chemistry analyses, detailed below. All filtrate water samples, filters and soil samples are cooled to 4 °C and transported immediately back to the laboratory in Wairakei where they are then stored as listed below until sent for analysis.
All chemistry analyses are performed at either the New Zealand Geothermal Analytical Laboratory (NZGAL) or the Extremophile Research Group (both at GNS Science), or the University of Waikato’s School of Science. Also all the metadata recorded on the tablet by the field table is uploaded to the database, where it instantly appears on the website.

Sometimes weather conditions, location of the hot spring or the nature of the sample can result in processing being performed in the laboratory. Many of the hot springs are geographically situated within a 20 km radius of GNS Science. Some of the samples are high in particulate matter and need time to settle before filtering.
The below table lists processing steps of samples and standard analytical methods used to measure the different variables.Sample Bottle |
Processing |
Storage |
Parameter |
Analytical Method |
Location |
|
2 L bottle |
Filtered |
-20 °C |
Microbial diversity |
DNA extraction & sequencing |
UWaikato |
|
500 mL bottle |
Raw |
NA |
Physical properties1 |
Multiparameter field meter |
On site |
|
500 mL bottle |
Filtered (0.22 µm) |
NA |
Fe2+ |
On site |
|
|
330 mL rubber-sealed bottle |
Raw |
4 °C |
H2S |
NZGAL |
|
|
330 mL rubber-sealed bottle |
Raw |
4 °C |
HCO3 |
Automated titration |
NZGAL |
|
330 mL rubber-sealed bottle |
Raw |
4 °C |
Cl- 2 |
Automated titration |
NZGAL |
|
50 mL tube |
Filtered |
4 °C |
SO42- |
NZGAL |
|
|
50 mL tube |
Filtered |
-20 °C |
NH4+, PO43-, NO2-, NO3- |
UWaikato |
|
|
50 mL tube |
Filtered |
4 °C |
Back up sample |
NA |
NA |
|
15 mL tube |
Filtered & acidified |
4 °C |
Elements3 |
UWaikato |
|
|
15 mL tube |
Filtered & alkalified |
4 °C |
As, Sb |
NZGAL |
|
|
50 mL syringe |
Filtered |
RT |
H2, CH4, CO |
Extremophile Research Group, GNS |
NA: not applicable, RT: room temperature
Notes:
- 1 Physical properties measured by the field meter are pH, oxidation-reduction potential (ORP or redox), conductivity, total dissolved solids, turbidity and dissolved oxygen.
- 2 Low levels of chloride are analysed using ion chromatography, also by the NZGAL.
- 3 Elements measured using ICP-MS are boron, lithium, sodium, magnesium, aluminium, silicon, sulphur, potassium, calcium, vanadium, chromium, manganese, iron, cobalt, nickel, copper, zinc, arsenic, selenium, bromine, rubidium, selenium, molybdenum, silver, cadmium, caesium, barium, mercury, thallium, lead and uranium.
References
Wilson, A.D. 1960. The micro-determination of ferrous iron in silicate minerals by a volumetric and a colorimetric method. Analyst, 85, 823-827.
Coming soon!
Sequencing
Microbial Diversity Assessment
The number of microorganisms in soil and in freshwater can be upward of one billion cells and ten million cells respectively (Whitman et al., 1998). It is therefore difficult to count and impossible to determine the identities of all the bacteria and archaea in a geothermal hotspring sample. To determine the diversity of the microorganisms inhabiting a spring, we extract the total microbial DNA content filtered from the sample. We then read and compare the DNA sequence of gene universal to all life. The order of the nucleotides in this gene can be used to catagorise the microorganism it was retrieved from into a taxonomic category. We use Next-Generation Sequencing (NGS) technology (Ion Torrent) to sequence large numbers of sequences from multiple samples in parallel

DNA extraction
Summary: DNA can be extracted, purified and concentrated from microorganisms using a combination of chemical and physical techniques. Prior to cell disruption, we block possible binding sites on any solids/sediments captured in the filtration process with sterile skim milk. We then disrupt the captured cells in the Sterivex filter using cetyl trimethylammonium bromide (CTAB), and then chemically remove the proteins, lipids and ribonucleic acids (RNA) using a phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol. The DNA is then concentrated via a magnetic bead-adsorption protocol.
- Cell disruption. The Sterivex filters are thawed on ice. 1.0 mL 0.8 % w/v skim milk powder solution is added first to the filters and incubated for 30 min at 65 °C. 0.4 mL of CTAB, and 0.2 mL PBS, and 0.1 mL SDS are then added and vortexed for 1 min. The filter is then placed in a 50 mL sterile capped plastic tube and incubated at 150 rpm 65 °C for 30 mins. The filter is then cooled on ice for a few minutes. The aqueous component is then pushed through the syringe and collected in a 2 mL tubes.
- DNA purification. Add equal volume (1:1) of chloroform: isoamyl alcohol (24:1) to each tube, vortex for 10 s, centrifuge at 12 400 rpm for 10 mins and transfer aqueous phase (usually top) to a new tube, avoid bottom layer. Add 300 µL chloroform: isoamyl (24:1) to the aqueous phase, mix tubes on Hula mixer for 20 mins. Centrifuge for 10 mins at 12 400 rpm and then collect aqueous phase into new tubes, avoiding the bottom layers.
- DNA concentration. Add equal volume of 100 % ethanol and 40 µL MoBio beads to the aqueous phase. Agitate every 30 s to 1-5 mins. Place samples on magnetic stand for 2 mins. Remove supernatant and wash beads with 500 µL of 100% ethanol, clean walls of tubes. Place tubes back on magnetic rack, remove all ethanol. Resuspend beads in 30 µL Tris (pH 8.0) by gentle pipetting and brief vortexing.
DNA amplification and preparation for Next Generation Sequencing

Summary: We determine the microbial diversity of each sample by sequencing (reading) the DNA code of a short gene that is possessed by all life called the small subunit of the ribosomal RNA (SSU rRNA). In bacteria and archaea, this is called the 16S rRNA gene and is approximately 1540 nucleotides long (in eukaryotes including humans it is called the 18S rRNA gene and is ~ 1870 nt). The total DNA content is extracted and the 16S rRNA genes of all resident bacteria and archaea is amplified using the Polymerase Chain Reaction (PCR) modified to facilitate NGS using the Ion Torrent . On average approximately 70,000 sequences (~270 bp in length) were read per sample.
- DNA amplification. We used the PCR to amplify the microbial community 16S rRNA genes using a universal bacterial/archaeal primer set F515 (5’-GTGCCAGCMGCCGCGGTAA-3’) and R806 (5’-GGACTACVSGGGTATCTAAT-3’) (Earth Microbiome Project [link: http://www.earthmicrobiome.org/emp-standard-protocols/16s/]). The forward primer contained an A-adaptor sequence, a barcode sequence 10-12 nt unique to each sample, a ‘GAT’ barcode adaptor sequence and the F515 sequencing primer. The reverse primer included a P1 sequence and R806 sequencing primer.
- PCR reactions were performed in 25 µL reactions, each containing 0.5 µL (10 µM concentration) of forward and reverse primers, 1.0 µL BSA (10 mg mL-1), 3.0 µL dNTPs (8 mM), 3.0 µL 10X PCR buffer, 3.0 µL MgCl2 (50 mM), and 2 µL soil community DNA at 0.5 ng/µL as a template. Prior to the addition of primers, Taq and template, the PCR master mix was treated with 18.6 µL ethidium monoazide bromide (1/200 dilution from 1mg mL-1 stock) per reaction, with a 1 min incubation in darkness on ice and 1 min of photoactivation using a lamp to remove trace DNA contaminants (Rueckert & Morgan, 2007). The PCR reagents were supplied by Invitrogen (Invitrogen, Carlsbad, CA) and the ethidium monoazide bromide was supplied by Norrie Biotech (Auckland, NZ). PCR was conducted using the following thermocycling parameters after the initial 3 min of 94 °C denaturation: 30 cycles (94 °C, 45 sec; 50 °C, 1 min; 72 °C, 1.5 min), followed by a final 10 min incubation at 72 °C.
-
 NGS preparation. The amplicons from triplicated PCR product pooling were purified using SPRIselect (Beckman Coulter Inc.) to remove small nucleotide fragments (left-hand selection), quality and concentration of the amplicons were verified and adjusted to 26 pM via HS Qubit 2.0 (Life Technologies) and 9100 BioAnalyser (Agilent Technologies) prior to pooling amplicons from all samples together for emulsion PCR. DNA sequencing was conducted at the Waikato DNA Sequencing Facility (University of Waikato, New Zealand) using Ion Torrent PGM (Life Technologies) with Ion 318v2 chip, using 400 bps chemistry.
NGS preparation. The amplicons from triplicated PCR product pooling were purified using SPRIselect (Beckman Coulter Inc.) to remove small nucleotide fragments (left-hand selection), quality and concentration of the amplicons were verified and adjusted to 26 pM via HS Qubit 2.0 (Life Technologies) and 9100 BioAnalyser (Agilent Technologies) prior to pooling amplicons from all samples together for emulsion PCR. DNA sequencing was conducted at the Waikato DNA Sequencing Facility (University of Waikato, New Zealand) using Ion Torrent PGM (Life Technologies) with Ion 318v2 chip, using 400 bps chemistry.
Post-Sequencing processing
Summary: The raw sequences (on average 70,000 reads per sample) are too many to curate by hand and are instead handled by customised bioinformatic pipelines. Firstly, the raw sequences are curated to remove erroneous reads (too long or too short) and any tagging DNA sequences that were added as part of the sequencing process removed. The sequences are then screened for known errors and trimmed to 250 bp in length. Similar sequences are then grouped (groups have a sequence similarity of 97 %) into groups called Operational Taxonomic Units (OTUs) which can, for simplicity, be considered synonymous with a taxonomic species classification, although this is not technically correct. The OTUs are then taxonomically ranked by comparing the OTU sequences with microbial taxonomic databases. The OTU taxonomies and abundances within a sample can be represented in a number of ways. It should be noted that for the purposes of this website OTUs with abundances of >5 reads per sample have not been presented graphically for cl
Technical Details:The raw reads in FASTQ format were processed using a custom pipeline based on Mothur (Schloss et al, 2009) and USEARCH (Edgar, 2013). Reads with lengths outside the 275-345 bp range were removed, as were reads with any error in the barcode and forward PCR primer sequences. The barcode and forward primer sequences were removed from the remaining reads, which were subjected to Q-score-based filtering to remove those with ≥3 expected errors. The reads were globally trimmed to 250 bp. After de-replication, unique reads were sorted based on their associated abundance. USEARCH was used to generate OTUs using centroid-based clustering (97% similarity threshold) and remove chimeras. OTUs were taxonomically assigned from their representative reads using the RDP Classifier in conjunction with RDP Release 11.2 (Wang et al, 2007).
References
Edgar, R.C. 2013. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods 10, 996-998.
Rueckert, A. and Morgan, H.W. 2007. Removal of contaminating DNA from polymerase chain reaction using ethidium monoazide. J Microbiol Meth, 68(3), 596-600.
Schloss, P.D., Westcott, S.L., Ryabin, T., Hall, J.R., Hartmann, M., Hollister, E.B., Lesniewski, R.A., Oakley, B.B., Parks, D.H., Robinson, C.J., Sahl, J.W., Stres, B., Thallinger, G.G., Van Horn, D.J., and Weber, C.F. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol, 75(23), 7537-7541.
Wang Q., Garrity, G.M., Tiedje, J.M. and Cole, J.R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol, 73(16), 5261-5267.
Whitman, W.B., Colem and, D.C. and Wiebe, W.J. 1998. Prokaryotes, the unseen majority. Proc Natl Acad Sci USA, 95(12), 6578-6583.
Health and Safety

Health, Safety and Environment
Please note here that sampling springs in geothermal environments is dangerous and should not be attempted. Ground surrounding hotsprings can be hot, unstable and undercut. Springs also often emit high concentrations of toxic and/or asphyxiating gases. If visiting a geothermal environment, do not leave formed tracks and do not approach springs or steaming soils. All people involved in this project have extensive experience in moving through and sampling geothermal ecosystems. In all cases, we attempted to minimise any impact of sampling on the surrounding environment.
Health and Safety Personal Protective Equipment (PPE)
For those interested, we have listed here the PPE and H&S measures we employ during sampling. We carry a H&S emergency bag which includes a satellite phone, locator beacon and laminated information sheets including emergency contacts, locations, recommended PPE, and hazards applicable to each site. We also have a comprehensive burns kit with a full body gel blanket, a 1st aid kit, collapsible bucket, water bladder, modified throwbag, gas monitor and GPS. We prefer to wear leather boots topped with neoprene putty’s (like gaters) over using gumboots. We also wear locally-made Cactus SuperTrousers to protect against vegetation and slow the infiltration of water/mud if an accident occurs.




